The image quality of ultrasound localization microscopy (ULM) images is driven by the ability to accurately detect and track the location of microbubbles (MBs) in vascular networks. This task becomes increasingly challenging in imaging environments with high MB concentrations and low signal-to-noise ratios, making it difficult to differentiate and localize individual MBs. Recent developments in deep learning (DL) have demonstrated significant improvements over conventional methods but depend on vast amounts of realistic training data with the corresponding ground truth labels, which are difficult to obtain. The alternative, simulated data, in turn, poses challenges in generalizability of the method. In this work, we present a hybrid pipeline for ULM that comprises data generation, localization, and tracking. It combines the current state-of-the-art, utilizing both conventional and DL techniques. We show that using this approach, we can create high-quality velocity maps while being able to generalize well across different domains.
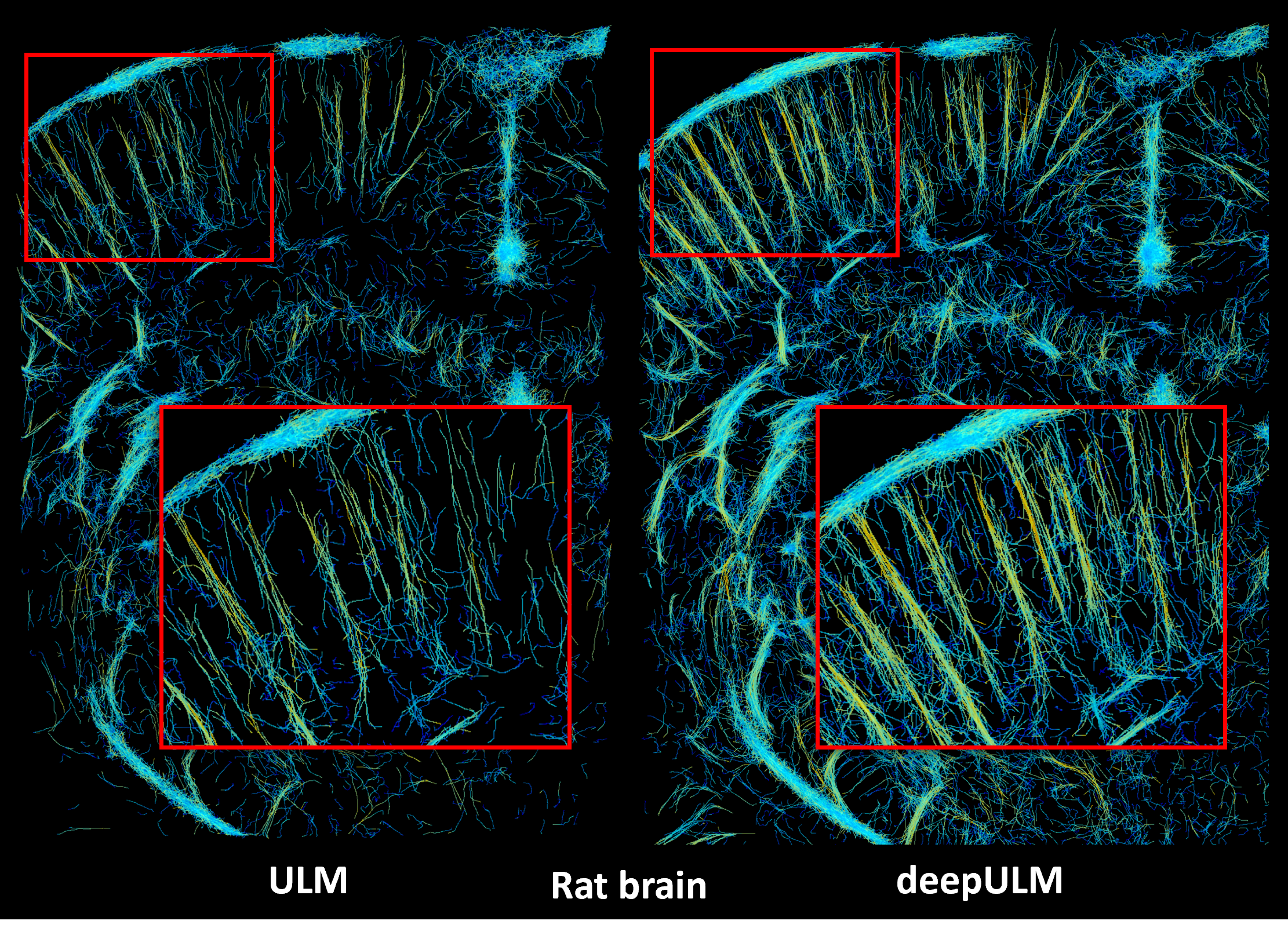
Results on the in-vivo rat brain data of the challenge. Comparing both ULM and deepULM images